Carboxysomes are small compartments inside photosynthetic bacteria where the machinery for capturing carbon dioxide is concentrated. You can see carboxysomes and their characteristic virus-like shape when you look at slices of these bacteria under an electron microscope:
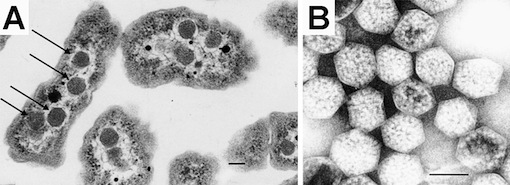
 Until recently, no one had looked at carboxysomes under the microscope in cells that were still alive. My labmates Dave and Bruno developed a way to label carboxysomes with fluorescent proteins and track them under a microscope as the cells grow, and their amazing paper in Science details some of the fascinating systems they discovered about how carboxysomes are controlled.
Until recently, no one had looked at carboxysomes under the microscope in cells that were still alive. My labmates Dave and Bruno developed a way to label carboxysomes with fluorescent proteins and track them under a microscope as the cells grow, and their amazing paper in Science details some of the fascinating systems they discovered about how carboxysomes are controlled.
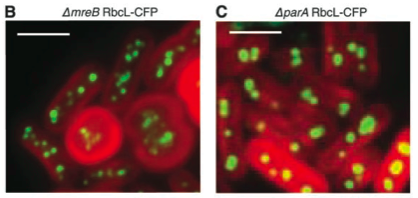 They noticed something very interesting right away: the carboxysomes in live cells are all lined up, evenly spaced, down the central axis of the rod-shaped bacteria. They hypothesized that there must be something holding the carboxysomes in place, preventing them from diffusing through the cytoplasm. All bacteria have a "skeleton," a mesh of proteins that maintains their shape, helps them divide, and can hold chromosomes and other cellular parts in place. When they deleted one of these mesh proteins out of the genome of the photosynthetic bacteria they saw that the cells would become rounder, not able to hold their shape as well, and that the carboxysomes weren't evenly spaced any more (figure B). When they knocked out a different skeleton-associated protein, parA, they saw that it seemed to exert special control over the carboxysomes. Deleting this gene allowed the cells to stay rod-shaped, but the carboxysomes weren't lined up anymore (figure C).
They noticed something very interesting right away: the carboxysomes in live cells are all lined up, evenly spaced, down the central axis of the rod-shaped bacteria. They hypothesized that there must be something holding the carboxysomes in place, preventing them from diffusing through the cytoplasm. All bacteria have a "skeleton," a mesh of proteins that maintains their shape, helps them divide, and can hold chromosomes and other cellular parts in place. When they deleted one of these mesh proteins out of the genome of the photosynthetic bacteria they saw that the cells would become rounder, not able to hold their shape as well, and that the carboxysomes weren't evenly spaced any more (figure B). When they knocked out a different skeleton-associated protein, parA, they saw that it seemed to exert special control over the carboxysomes. Deleting this gene allowed the cells to stay rod-shaped, but the carboxysomes weren't lined up anymore (figure C).
In the mutants without parA and no even carboxysome spacing, sometimes when a cell divided, one of its daughter cells wouldn't get any carboxysomes. Without the machinery to capture carbon dioxide, the cell grew much slower until it was able to get enough protein together to make a new carboxysome. In the video below you can see this happening. The red arrow points to a cell that gets no carboxysomes after division, and the white arrow points at its sister cell that got them all. The empty cell doesn't divide again until it forms carboxysomes (the green dots), while the cell that got the carboxysomes has already divided by that time. This shows why the cell would invest so much energy holding the carboxysomes in place; without even spacing a certain number of cells wouldn't be able to grow at the optimal speed, decreasing the fitness of the whole population.
When they fluorescently tagged parA in wild type cells, they saw something amazing: this "skeleton" protein isn't just a static structure that the carboxysomes cling too, but an oscillating wave, ping-ponging back and forth down the length of the bacteria. As the wave moves through the cell parA makes sure that the carboxysomes are evenly spaced along the whole axis. In other species of bacteria, this skeleton wave can control the even spacing of genetic material along the length of the cell or keeps proteins associated with the tips of rod-shaped bacteria where they belong. You can see the wave traveling through the cell in the second video, where the carboxysomes are labeled in red and the wave protein in green.
With a deeper understanding of the cell biology of the carboxysomes and how they are controlled in the cell, as well as the genetic tools that Bruno and Dave developed for putting the fluorescent proteins into the photosynthetic bacteria, synthetic biologists in our lab and others will be better poised to engineer the carboxysomes for any number of synthetic purposes, to design novel bacterial micro-factories.

 Notes, thoughts, and news on synthetic biology by
Notes, thoughts, and news on synthetic biology by 


Comments
Very well explained.
Posted by: Art | March 14, 2010 2:53 AM
Excellent blog, i am jealous of you students because you have great writing skills, keep it up, Greetings form Mexico.
Posted by: MSemán | March 16, 2010 2:16 AM