Well, I really was very much enthused, inspired and uplifted by the many kind and supportive comments so many of you added to the previous article. Thank you all. So enthused, in fact, that I couldn't help myself, and took time out of lunch breaks and so on to produce 'ticking over' material for Tet Zoo.
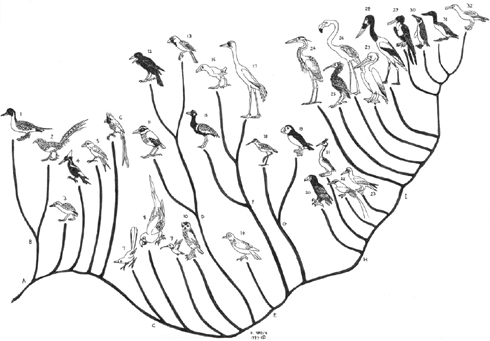
As some of you know, I'm hard at work on a major project concerning birds at the moment (and, to the people who made speculations that such project might be lucrative... let me assure you that none of the science writing I do can be described as 'lucrative' - no money is involved in this project, for example. Do people realise how much work scientists do for free?). In view of this, I dug out the old image you see here (it was produced in 1997). What does it mean? Here's an excerpt from the text I'm currently working on...

What appeared to be an empirical approach to the study of the avian tree came to the fore during the 1980s and 90s when Charles Sibley and colleagues used DNA-DNA hybridization to analyse avian relationships. Sibley and Ahlquist (1990) produced a mostly resolved phylogeny, dubbed the 'Tapestry', for over 1100 species.
They supported a basal divergence between Eoaves (ratites and tinamous) and Neoaves, and broke the latter down into six major assemblages: Galloanserae (gamebirds and waterfowl), Turnicae (buttonquails), Picae (woodpeckers and kin), Coraciae (hornbills, trogons, rollers and kin), Coliae (collies) and Passerae (everything else, from cuckoos, parrots and pigeons to cranes, waders, raptors, herons, pelicans and passerines). While Sibley and Ahlquist's (1990) suggestions did much to inspire new work, their DNA-DNA hybridization technique was entirely phenetic and their conclusions were not always supported by their data (Harshman 1994, 2007).
There's a lot more that could be said, but this was mostly an excuse to re-use the old 'Tapestry' picture used at top. Feel free to discuss among yourselves. Lots more on bird phylogeny to appear here in time.
Refs - -
Harshman, J. 1994. Reweaving the Tapestry: what can we learn from Sibley and Ahlquist (1990)? Auk 111, 377-388.
Harshman, J. 2007. Classification and phylogeny of birds. In Jamieson, B. G. M. (ed) Reproductive Biology and Phylogeny of Birds. Science Publishers, Inc. (Enfield, NH), pp. 1-35.
Sibley, C. G. & Ahlquist, J. A. 1990. Phylogeny and Classification of Birds. New Haven: Yale University Press.

 With six years of phd work on theropod dinosaurs behind him, Darren Naish mostly spends long hours in the library, hunched over his laptop. But he gets out sometimes, and picks up litter and pursues exotic lizards across the British countryside, aiming all the while to publish his technical work on obscure Cretaceous dinosaurs. He also messes around with pterosaurs, swimming giraffes, British big cats and stuff like that. He has given up on the stupid idea of being a dedicated academic and ekes out a living as a technical consultant, editor and author. He can be contacted intermittently at eotyrannus (at) gmail dot com. For more biographical info go
With six years of phd work on theropod dinosaurs behind him, Darren Naish mostly spends long hours in the library, hunched over his laptop. But he gets out sometimes, and picks up litter and pursues exotic lizards across the British countryside, aiming all the while to publish his technical work on obscure Cretaceous dinosaurs. He also messes around with pterosaurs, swimming giraffes, British big cats and stuff like that. He has given up on the stupid idea of being a dedicated academic and ekes out a living as a technical consultant, editor and author. He can be contacted intermittently at eotyrannus (at) gmail dot com. For more biographical info go 












Comments
Yay! for "ticking over" material.
I hadn't commented yet on the "Death of..." post, but add me to the number of people who were alarmed at the headline and profoundly relieved that this only meant time out and not forever. Totally get needing the time...but it would be heartbreaking to lose this blog for good.
Re. the tapestry -- love the picture. Shame their conclusions weren't as good as they could be.
Posted by: Luna_the_cat | February 12, 2010 7:24 AM
Yay! Many thanks for that!
I don't know what it is but I have my suspicions...
Is that an ivory-billed woodpecker I see in the bird tree?
Posted by: Dartian | February 12, 2010 7:31 AM
Yup, that's an Ivory-billed. Now identify all the others :)
Posted by: Darren Naish | February 12, 2010 7:36 AM
Yes, Sibley and Ahlquist's reach certainly exceeded their grasp, yet their tapestry still has a lot of support from less phylogenetic ornithologists and among amateur birders.
By the way, I also wanted to extend my compliments to you for all your hard work on this blog, Darren. It's one of several biology blogs I check on frequently. And for none of these favorite blogs do I feel cheated if there is no new material. I can wait. There is plenty for me to read and keep up with in science, but whenever my favorite blogger adds an interesting piece I will be there to devour it.
Posted by: Diego | February 12, 2010 8:25 AM
Only scientists and perhaps their spouses do, barely even their parents. Other people would never guess. One word: page charges.
Well, I wouldn't even say they produced a phylogeny. They produced a phenetic hypothesis and sold it as a phylogenetic hypothesis…
Posted by: David Marjanović | February 12, 2010 8:36 AM
take as long of a break as you need whenever you need..you owe nothing..thanks for everything..we`ll be here..
sluggo
Posted by: sluggo | February 12, 2010 9:06 AM
We are all glad to see a post again, I think you may just post with less frequency as you finish your project.
"Do people realise how much work scientists do for free?"
that is an perennial issue regarding science.
Posted by: Jorge W. Moreno-Bernal | February 12, 2010 9:17 AM
I can't identify all, but here are a few (in no particular order) that I'm fairly confident I have down to correct species:
pintail duck
Lady Amherst's pheasant
saddle-billed stork
grey heron
northern bald ibis
whooping crane
great crested grebe
Atlantic puffin
great northern loon
bateleur
emperor penguin
greater flamingo
great white pelican
The bustard is an Afrotis species, and the mousebird is some Colius (not Urocolius) species. The frigatebird, the tropicbird and the pitta, at least, are probably identifiable to species level too (though not by me).
Posted by: Dartian | February 12, 2010 9:53 AM
I like that drawing for the alertness in the birds. In illos like that, they usually look very static, but not in that one.
Posted by: Mike from Ottawa | February 12, 2010 10:02 AM
I see a The Saddle-billed Stork (Ephippiorhynchus senegalensis)towards the end
Posted by: gorilla | February 12, 2010 10:11 AM
I have to second Diego's comment (#4) above -- good job on managing such a great science blog!
I also wanted to mention a few other refs that may be of interest:
Shannon J. Hackett, et al. A Phylogenomic Study of Birds Reveals Their Evolutionary History Science 320, 1763 (2008); DOI: 10.1126/science.1157704
For some recent work on Parulidae (new world warblers): http://www.birds.cornell.edu/evb/Irby.htm
As mentioned in the page linked above, a number of groups have been coordinating efforts to refine relationships within Emberizidae. I'm not sure how much of that has been published yet -- it might still be "work in progress".
Posted by: ObSciGuy | February 12, 2010 11:44 AM
Ticking over's good! Take your time, we'll be here.
Posted by: pomposa | February 12, 2010 1:27 PM
Dartian: I'm pretty confident that the parrot is a scarlet macaw. Although I guess it could be a green-winged macaw, depending if the clear face is an artifact of the resolution.
Posted by: Sebastian Marquez | February 12, 2010 4:09 PM
I also hadn't commented on the previous thread but like so many others was momentarily aghast that the 'death' might be permanent. Glad to see that you can't stay away Darren. and hope that you are able to continue educating your readers as well as earning a crust and looking after your family!
I have always been a big fan of your artwork and get the feeling you probably have lots of these gems hanging around which would make excellent images for the blog.
Identifications I can try to make to add to Dartian's:
No 10: Burrowing Owl
No 11: Indian Pitta?
No 22: Not Red Billed Tropicbird (as no back barring) so must be Red-tailed or White-tailed
No 23: Masked Booby?
Posted by: RStretton | February 12, 2010 4:46 PM
20 years since that book was published ... my, has time flown?
Posted by: Rob | February 12, 2010 8:21 PM
Darren,
Nice image sample of the "Tapestry". When I was a grad student, I had the full, species-level Tapestry taped around three walls of my office. And it was mighty impressive.
And I'd like to complain about you and David Marjanovic using "phenetic" as a swear word. There's nothing wrong with phenetic methods, per se. Like any methods, they produce good estimates of phylogeny to the extent that the data don't violate the assumptions of the method. UPGMA will give you a decent phylogeny if there is a very good molecular clock. Sibley & Ahlquist had some problems, not least of which was the failure of that molecular clock in quite a few spots. Then again, many of their most interesting results, particularly within passerines, have held up. And one other triumph selected at random: their charadriiform phylogeny was spot on.
One quibble: Eoaves was originally named by S&A; to include paleognaths and Galloanserae, a novel -- and incorrect -- clade. And Neoaves was Eoaves' sister group, all other gbirds. When that didn't pan out, they tried to redefine the terms to include Galloanserae within Neoaves. Fortunately, everyone ignored them (since the names would have become junior synonyms of Palaeognathae and Neognathae). Eoaves died a deserved death, but Neoaves remains.
Posted by: John Harshman | February 12, 2010 10:31 PM
Well, yeah. The assumptions of the method of using phenetics to do phylogenetics are that there's little enough homoplasy in the data. And that's among the very possibilities that a phylogenetic analysis is supposed to test. Just assuming them is a spectacularly bad idea, as lots of empirical examples show, for instance every single case of long-branch attraction.
Indeed – there just almost never is a halfway decent molecular clock.
I think we can agree that precladistic morphological phylogenetics (an art rather than a science) was occasionally even worse than using molecular phenetics as phylogenetics. :-)
Long-branch attraction of the outgroups to the long stem of Neoaves.
Posted by: David Marjanović | February 13, 2010 7:37 AM
I still disagree. Long branch attraction is hardly a special problem of phenetic methods. Let's remember that it was first demonstrated using parsimony. Nor do all distance methods assume that homoplasy is rare; depends on the distance measure -- everything from Jukes-Cantor distance on up attempt to correct for homoplasy.
In some circles it's enough to sniff "feh, phenetic" in order to ignore an entire analysis. That's convenient, but it's lazy. Case in point: I tell you that S&A;'s phylogeny of charadriiforms was exactly right, and you sniff that precladistic morphological phylogenetics was even worse. Even worse than exactly right?
I will point out that there are phenetic (=distance?) methods that don't assume a clock, and they solve many of UPGMA's problems using the same data. Even so, within some clades, like passerines, there seems to be a good enough clock, and in others, like anseriforms, there's only one problematic node even using UPGMA. (The best published study of anseriform phylogeny is still Madsen et al. 1988, using UPGMA.)
And the problem with Eoaves wasn't exactly long-branch attraction. It was extrapolation of distance measures long past their useful range. UPGMA in S&A;'s results was actually more vulnerable to short-branch attraction.
Posted by: John Harshman | February 13, 2010 9:35 AM
Cool old stuff...
... but what is this bird project?
Posted by: Jerzy | February 13, 2010 11:10 AM
No, but they're more susceptible to it than phylogenetic methods, even parsimony.
To the contrary. The only good reason there ever was for using phenetic instead of phylogenetic methods for doing phylogenetics is that the phenetic ones are a lot faster. But with today's computers, calculation time hardly ever is an issue anymore; parsimony is fast enough, and maximum likelihood and Bayesian inference are usually feasible, too.
Yes. Phenetics measures the similarity ( = 1 - distance); phylogenetics counts the shared derived character states (whether transformed into likelihoods or not).
Similarity can be synapomorphic, symplesiomorphic, or homoplastic. Phylogenetic methods sometimes fail to distinguish these correctly; phenetic ones don't even try.
I was making a general point, not one specific to this example.
Why?
That's the same thing – it means that the long branches wander off together, so that the short branches are left.
Posted by: David Marjanović | February 13, 2010 1:43 PM
Several studies have found this not to be the case, particularly when the distances used match the model under which the characters evolved. What do you have on that? And by the way, I object to the claim that phenetic methods aren't phylogenetic.
Likelihood doesn't count shared derived character states. It counts all states, and applies the same computations to all.
Because it gets the phylogeny closest to right. Unfortunately I can't show you my criteria for "right", since my study (which I hope we will all agree is right by definition) isn't yet published.
Finally, I deny that long-branch and short-branch attraction are at all the same thing. They may have the same result, but the reasons are wholly different. UPGMA is subject to SBA simply because it clusters the most similar taxa; all you need for a mistake is enough variation in evolutionary rate. LBA, on the other hand, requires non-additive distances, i.e. that the measured distances are increasingly underestimates of the true distance as true distance increases.
Posted by: John Harshman | February 13, 2010 6:59 PM
Oh, and by the way, just what is your mysterious bird project? Or is it secret?
Posted by: John Harshman | February 13, 2010 7:02 PM
Thanks to all for comments. Interesting stuff on phenetic techniques, thanks John. What you say has made me re-think my opinion on this subject.
Birds in 'Tapestry' pic are meant to be (from left to right): Pintail, Lady Amherst's pheasant, Andalusian hemipode, Ivory-billed woodpecker, Lilac-breasted roller, Speckled mousebird, Coccyzus cuckoo, generic macaw, generic hummingbird, Burrowing owl, Banded pitta, Australian raven, Desert sparrow, Rufous turtle dove, Black korhaan, Purple gallinule, Whooping crane, wader (Redshank?), Atlantic puffin, Bateleur, Great crested grebe, Red-tailed tropicbird, Masked booby, Purple heron, Bald ibis, Greater flamingo, White pelican, Saddle-billed stork, frigate bird, Emperor penguin, Great northern diver and Wandering albatross.
Posted by: Darren Naish | February 14, 2010 6:54 AM
I'll try to look for references tomorrow.
I can only repeat: "Similarity can be synapomorphic, symplesiomorphic, or homoplastic. Phylogenetic methods sometimes fail to distinguish these correctly; phenetic ones don't even try." They have no qualms about clustering taxa based on their symplesiomorphies if those are just numerous enough.
Posted by: David Marjanović | February 14, 2010 8:18 AM
David,
Your criticism is not of phenetic methods, but of phenetic methods that assume a molecular clock. Those are the only ones that would cluster based on symplesiomorphies. (That's the short branch attraction I was talking about). Other methods, like least squares, do no such thing.
I can only repeat: any method is phylogenetic if it's intended to estimate phylogeny, and it will correctly estimate that phylogeny to the extent that its assumptions fit the data. Parsimony will perform better than UPGMA under some circumstances, but UPGMA will perform better than parsimony under others. I agree that the former case is more common than the latter, though of course parsimony is inapplicable to DNA hybridization. And UPGMA is hardly the only phenetic method. Cases in which, say, neighbor-joining outperforms parsimony are more common, and fairly easy to simulate.
Posted by: John Harshman | February 14, 2010 9:42 AM
I have a question about phenetics. We always hear it's inferior to cladistics because the latter only uses shared DERIVED characters, while phenetics uses any shared metric. But the only way PAUP knows a character is derived is via comparison to the outgroup. And it only holds true for a part of the tree anyway. What's a symplesiomorphy among Theropoda basally could be a synapomorphy within Tyrannosauroidea. Also, more and more authors are recognizing that it's best to split characters into multiple states, or even have them fully quantified as TNT allows. This is similar to many traditional phenetic analyses. So really, the only difference is that cladistic analyses root their trees automatically based on the outgroup, while you'd have to manually root a phenetic tree, no?
Posted by: Mickey Mortimer | February 15, 2010 1:57 AM
No. Even if you fix the root in advance, phenetic analyses can cluster taxa based, ultimately, on character states that parsimony would optimize as symplesiomorphies.
Phenetics doesn't look at the individual characters. It transforms the character matrix into a distance matrix and then works on the distance matrix. Cladistics... parsimony at least transforms the character matrix into a difference matrix and then works on the difference matrix, which shows which OTUs differ in which characters – something that's ignored in phenetics, where the sum total of the distance between two OTUs is used instead.
Models of evolution can be applied to both. In the case of parsimony, you get max. likelihood or, AFAIK, Bayesian inference. In the case of phenetics, you get neighbor-joining, AFAIK.
Test if the <pre> tag works:
test test testPosted by: David Marjanović | February 15, 2010 5:51 AM
Test utterly failed. It works on Pharyngula...
Posted by: David Marjanović | February 15, 2010 5:54 AM